Image Classification using Pretrained VGG Activation Maps as Features for SVM and FCNN models
This post consists of:
- Import Libraries
- Loading Data
- Pretrained Feature Extraction
- Feature Reduction
- PCA
- 2D PCA
- Visualization
- 3D PCA
- Visualization
- t-SNE
- 2D t-SNE
- Visualization
- 3D t-SNE
- Visualization
- PCA
- Train Test Split
- Train Models
- PCA
- SVC Linear - PCA 2D
- SVC Linear - PCA 3D
- SVC RBF - PCA 2D
- SVC RBF - PCA 3D
- t-SNE
- SVC Linear - t-SNE 2D
- SVC Linear - t-SNE 3D
- SVC RBF - t-SNE 2D
- SVC RBF - t-SNE 3D
- Fully Connected
- PCA
1 Import Libraries
%tensorflow_version 1.x
from __future__ import print_function
import pickle
import seaborn as sns
import matplotlib.pylab as plt
import PIL
from mpl_toolkits.mplot3d import Axes3D
import pandas as pd
import numpy as np
from sklearn.svm import SVC, LinearSVC
from sklearn.svm import base
from sklearn.metrics import confusion_matrix
from sklearn.decomposition import PCA
from sklearn.manifold import TSNE
import keras
from keras import backend as K
from keras.preprocessing.image import ImageDataGenerator
from keras.optimizers import Adam
from keras.callbacks import EarlyStopping, ReduceLROnPlateau, ModelCheckpoint, LearningRateScheduler
from keras.layers import Dense, Conv2D, BatchNormalization, Activation
from keras.layers import AveragePooling2D, Input, Flatten
from keras.regularizers import l2
from keras.models import Model
from keras.utils import to_categorical
from keras.applications.vgg19 import VGG19
/usr/local/lib/python3.6/dist-packages/sklearn/utils/deprecation.py:144: FutureWarning: The sklearn.svm.base module is deprecated in version 0.22 and will be removed in version 0.24. The corresponding classes / functions should instead be imported from sklearn.svm. Anything that cannot be imported from sklearn.svm is now part of the private API.
warnings.warn(message, FutureWarning)
Using TensorFlow backend.
2 Loading Data
You need to put your data in the following structure:
data/train/
/class1
/class2
/...
data/test/
/class1
/class2
/...
from google.colab import drive
drive.mount('/content/drive')
!unrar x drive/My\ Drive/IUST-PR/HW3/data.rar /content
# loading data
batch_size = 64
n_classes = 3
img_size = (224, 224)
datagen = ImageDataGenerator()
train_iterator = datagen.flow_from_directory('data/train/', class_mode='categorical',
batch_size=batch_size, target_size=img_size, shuffle=True)
# only for NN model
test_iterator = datagen.flow_from_directory('data/test/', class_mode='categorical',
batch_size=batch_size, target_size=img_size, shuffle=True)
n = 541
x = np.empty((n, img_size[0], img_size[1], n_classes))
y = np.empty((n, n_classes))
for idx in range(len(train_iterator)):
batchx = train_iterator[idx][0]
batchy = train_iterator[idx][1]
if batchx.shape[0] < batch_size:
x[idx*batch_size:] = batchx
y[idx*batch_size:] = batchy
else:
x[idx*batch_size:(idx+1)*batch_size] = batchx
y[idx*batch_size:(idx+1)*batch_size] = batchy
n_test = 31
y_test = np.empty((n_test, n_classes))
for idx in range(len(test_iterator)):
batchy = test_iterator[idx][1]
if batchx.shape[0] < batch_size:
y_test[idx*batch_size:] = batchy
else:
y_test[idx*batch_size:(idx+1)*batch_size] = batchy
print(x.shape)
print(y.shape)
print(y_test.shape)
Found 541 images belonging to 3 classes.
Found 31 images belonging to 3 classes.
(541, 224, 224, 3)
(541, 3)
(31, 3)
3 Pretrained Feature Extraction
When a CNN model is trained on a large dataset that can represent images similar to our custom dataset, has features that are common in first layers and more specific in the last layers. So we remove fully connected layers and use convolutional layers as the features extracted from ImageNet dataset where it is the superset of our dataset. These features even though are in low dimensional manner w.r.t. input images, they contain much more usefull information about images.
# feature extraction
model = VGG19(include_top=False, weights='imagenet', input_tensor=None, input_shape=(224, 224, 3), classes=1000)
features = model.predict_generator(train_iterator, steps=9)
features_test = model.predict_generator(test_iterator, steps=1)
print('NN data train shape:', features.shape)
print('NN data test shape:', features_test.shape)
NN data train shape: (541, 7, 7, 512)
NN data test shape: (31, 7, 7, 512)
4 Feature Reduction
In this step, we have 77512 features to feed to our SVM models to train. But we want to reduce this info and use only 2 and 3 feature that represent our whole features.
So we use PCA and t-SNE to reduce to 2 and 3 features.
- PCA
- 2D PCA
- Visualization
- 3D PCA
- Visualization
- t-SNE
- 2D t-SNE
- Visualization
- 3D t-SNE
- Visualization
4.A PCA
- 2D PCA
- Visualization
- 3D PCA
- Visualization
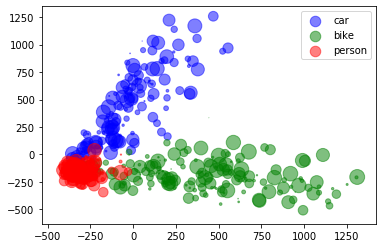
4.A.a 2D PCA
# 2D PCA
pca_2d = PCA(n_components=2)
pca_2d.fit(features.reshape(len(features), -1))
pca_2d_features = pca_2d.transform(features.reshape(len(features), -1))
print(pca_2d_features.shape)
(541, 2)
4.A.b Visualization
colors = np.argmax(y, axis=1)
fig, ax = plt.subplots()
x_ = pca_2d_features[:,0]
y_ = pca_2d_features[:,1]
legend_ = ['blue', 'green', 'red']
for idx, color in enumerate(['car', 'bike', 'person']):
area = (15 * np.random.rand(len(x)))**2
print(x_[colors==idx].shape, y_[colors==idx].shape, colors[colors==idx].shape)
ax.scatter(x_[colors==idx], y_[colors==idx], s=area, c=legend_[idx], alpha=0.5, label=color)
ax.legend()
plt.show()
(165,) (165,) (165,)
(169,) (169,) (169,)
(207,) (207,) (207,)
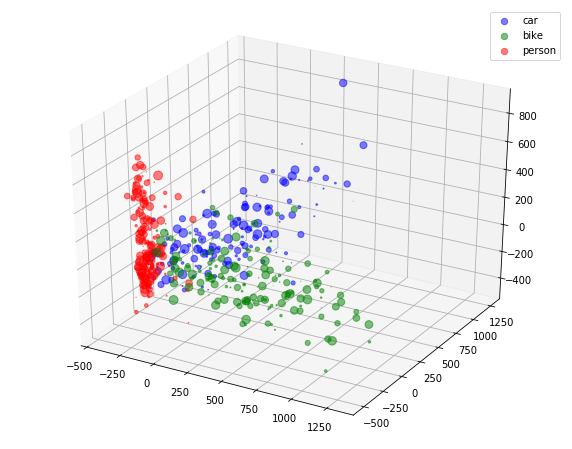
4.A.c 3D PCA
# 3D PCA
pca_3d = PCA(n_components=3)
pca_3d.fit(features.reshape(len(features), -1))
pca_3d_features = pca_3d.transform(features.reshape(len(features), -1))
print(pca_3d_features.shape)
(541, 3)
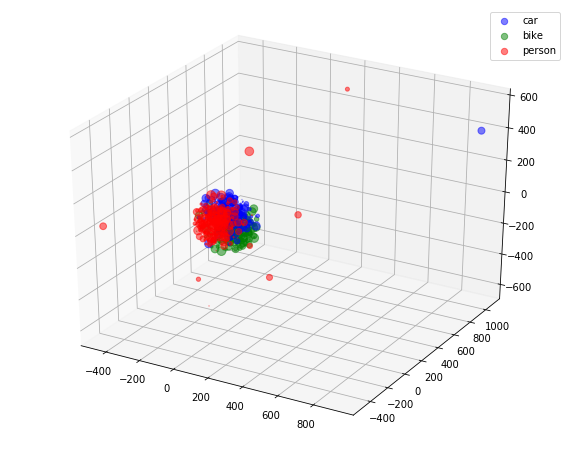
4.A.d Visualization
colors = np.argmax(y, axis=1)
fig = plt.figure(figsize=(10, 8))
ax = fig.add_subplot(111, projection='3d')
x_ = pca_3d_features[:, 0]
y_ = pca_3d_features[:, 1]
z_ = pca_3d_features[:, 2]
legend_ = ['blue', 'green', 'red']
for idx, color in enumerate(['car', 'bike', 'person']):
area = (9 * np.random.rand(len(x)))**2
print(x_[colors==idx].shape, y_[colors==idx].shape, colors[colors==idx].shape)
ax.scatter(x_[colors==idx], y_[colors==idx], z_[colors==idx], s=area, c=legend_[idx], alpha=0.5, label=color)
ax.legend()
plt.show()
(165,) (165,) (165,)
(169,) (169,) (169,)
(207,) (207,) (207,)
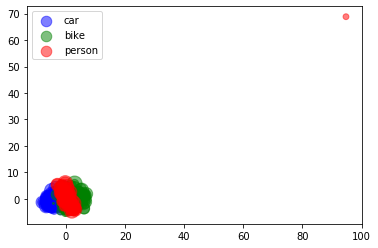
4.B t-SNE
- 2D t-SNE
- Visualization
- 3D t-SNE
- Visualization
4.B.a 2D t-SNE
# TSNE 2D
tsne_2d = TSNE(n_components=2, perplexity=30, learning_rate=200)
tsne_2d_features = tsne_2d.fit_transform(features.reshape(len(features), -1))
print(tsne_2d_features.shape)
(541, 2)
4.B.b Visualization
colors = np.argmax(y, axis=1)
fig, ax = plt.subplots()
x_ = tsne_2d_features[:,0]
y_ = tsne_2d_features[:,1]
legend_ = ['blue', 'green', 'red']
for idx, color in enumerate(['car', 'bike', 'person']):
area = (15 * np.random.rand(len(x)))**2
print(x_[colors==idx].shape, y_[colors==idx].shape, colors[colors==idx].shape)
ax.scatter(x_[colors==idx], y_[colors==idx], s=area, c=legend_[idx], alpha=0.5, label=color)
ax.legend()
plt.show()
(165,) (165,) (165,)
(169,) (169,) (169,)
(207,) (207,) (207,)
4.B.c 3D t-SNE
# TSNE 3D
tsne_3d = TSNE(n_components=3, perplexity=30, learning_rate=200)
tsne_3d_features = tsne_3d.fit_transform(features.reshape(len(features), -1))
print(tsne_3d_features.shape)
(541, 3)
4.B.d Visualization
colors = np.argmax(y, axis=1)
fig = plt.figure(figsize=(10, 8))
ax = fig.add_subplot(111, projection='3d')
x_ = tsne_3d_features[:, 0]
y_ = tsne_3d_features[:, 1]
z_ = tsne_3d_features[:, 2]
legend_ = ['blue', 'green', 'red']
for idx, color in enumerate(['car', 'bike', 'person']):
area = (9 * np.random.rand(len(x)))**2
print(x_[colors==idx].shape, y_[colors==idx].shape, colors[colors==idx].shape)
ax.scatter(x_[colors==idx], y_[colors==idx], z_[colors==idx], s=area, c=legend_[idx], alpha=0.5, label=color)
ax.legend()
plt.show()
(165,) (165,) (165,)
(169,) (169,) (169,)
(207,) (207,) (207,)
5 Train Test Split
For validating our trained model we need to separate a small proportion of our data to propose to our trained model where those have not been seen at all.
# train test split
from sklearn.model_selection import train_test_split
pca_2d_x_train, pca_2d_x_test, pca_2d_y_train, pca_2d_y_test = train_test_split(pca_2d_features,
np.argmax(y, axis=1),
test_size=0.1)
pca_3d_x_train, pca_3d_x_test, pca_3d_y_train, pca_3d_y_test = train_test_split(pca_3d_features,
np.argmax(y, axis=1),
test_size=0.1)
tsne_2d_x_train, tsne_2d_x_test, tsne_2d_y_train, tsne_2d_y_test = train_test_split(tsne_2d_features,
np.argmax(y, axis=1),
test_size=0.1)
tsne_3d_x_train, tsne_3d_x_test, tsne_3d_y_train, tsne_3d_y_test = train_test_split(tsne_3d_features,
np.argmax(y, axis=1),
test_size=0.1)
print('PCA 2D features train size:',pca_2d_x_train.shape, '------','PCA 2D features test size', pca_2d_x_test.shape)
print('PCA 3D features train size:',pca_3d_x_train.shape, '------','PCA 3D features test size', pca_3d_x_test.shape)
print('TSNE 2D features train size:',tsne_2d_x_train.shape, '------','TSNE 2D features test size', tsne_2d_x_test.shape)
print('TSNE 3D features train size:',tsne_3d_x_train.shape, '------','TSNE 3D features test size', tsne_3d_x_test.shape)
PCA 2D features train size: (486, 2) ------ PCA 2D features test size (55, 2)
PCA 3D features train size: (486, 3) ------ PCA 3D features test size (55, 3)
TSNE 2D features train size: (486, 2) ------ TSNE 2D features test size (55, 2)
TSNE 3D features train size: (486, 3) ------ TSNE 3D features test size (55, 3)
6 Train Models
- PCA 1. SVC Linear - PCA 2D 2. SVC Linear - PCA 3D 3. SVC RBF - PCA 2D 4. SVC RBF - PCA 3D
- t-SNE
- SVC Linear - t-SNE 2D
- SVC Linear - t-SNE 3D
- SVC RBF - t-SNE 2D
- SVC RBF - t-SNE 3D
- Fully Connected
# SVM
class SVM_MODELS:
"""
Enum of possible models
"""
SVC = 1
LINEAR_SVC = 2
# %% functions
def build_svm(model_type: int, x: np.ndarray, y: np.ndarray, svc_kernel: str = None, save: bool = True,
path: str = None, **kwargs) -> base:
"""
Trains a SVM model
:param model_type: The kernel of SVM model (see `SVM_MODELS` class)
:param svc_kernel: The possible kernels for `SVC` model (It must be one of ‘linear’, ‘poly’, ‘rbf’, ‘sigmoid’)
It will be ignored if `model_type = LINEAR_SVC`
:param x: Features in form of numpy ndarray
:param y: Labels in form of numpy ndarray
:param save: Whether save trained model on disc or not
:param path: Path to save fitted model
:param kwargs: A dictionary of other optional arguments of models in format of {'arg_name': value} (ex: {'C': 2})
:return:A trained VSM model
"""
if model_type == SVM_MODELS.SVC:
model = SVC(kernel=svc_kernel, **kwargs)
elif model_type == SVM_MODELS.LINEAR_SVC:
model = LinearSVC(**kwargs)
else:
raise Exception('Model {} is not valid'.format(model_type))
model.fit(x, y)
if save:
if path is None:
path = ''
pickle.dump(model, open(path + model.__module__ + '.model', 'wb'))
return model
6.A PCA
- SVC Linear - PCA 2D
- SVC Linear - PCA 3D
- SVC RBF - PCA 2D
- SVC RBF - PCA 3D
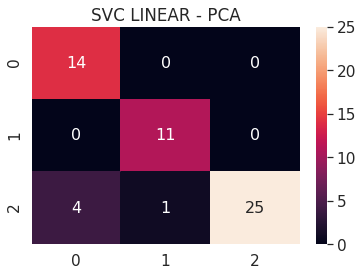
6.A.a SVC Linear - PCA 2D
# SVC LINEAR PCA 2D
pca_2d_svc_linear = build_svm(model_type=SVM_MODELS.LINEAR_SVC, x=pca_2d_x_train, y=pca_2d_y_train,
max_iter=200000, path='models/pca_2d_', multi_class='ovr')
print('PCA 2D - SVC LINEAR - Test accuracy', pca_2d_svc_linear.score(pca_2d_x_test, pca_2d_y_test))
cm = confusion_matrix(pca_2d_svc_linear.predict(pca_2d_x_test), pca_2d_y_test)
df_cm = pd.DataFrame(cm, range(n_classes), range(n_classes))
sns.set(font_scale=1.4)
sns.heatmap(df_cm, annot=True, annot_kws={"size": 16})
plt.title('SVC LINEAR - PCA')
plt.show()
/usr/local/lib/python3.6/dist-packages/sklearn/svm/_base.py:947: ConvergenceWarning: Liblinear failed to converge, increase the number of iterations.
"the number of iterations.", ConvergenceWarning)
PCA 2D - SVC LINEAR - Test accuracy 0.9090909090909091
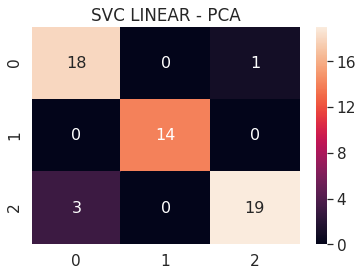
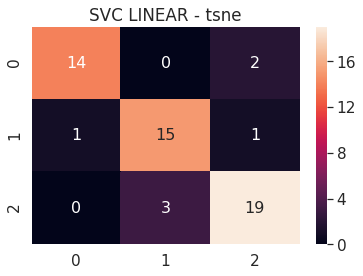
6.A.b SVC Linear - PCA 3D
# SVC LINEAR PCA 3D
pca_3d_svc_linear = build_svm(model_type=SVM_MODELS.LINEAR_SVC, x=pca_3d_x_train, y=pca_3d_y_train,
max_iter=200000, path='models/pca_3d_', multi_class='ovr')
print('PCA 3D - SVC LINEAR - Test accuracy', pca_3d_svc_linear.score(pca_3d_x_test, pca_3d_y_test))
cm = confusion_matrix(pca_3d_svc_linear.predict(pca_3d_x_test), pca_3d_y_test)
df_cm = pd.DataFrame(cm, range(n_classes), range(n_classes))
sns.set(font_scale=1.4)
sns.heatmap(df_cm, annot=True, annot_kws={"size": 16})
plt.title('SVC LINEAR - PCA')
plt.show()
/usr/local/lib/python3.6/dist-packages/sklearn/svm/_base.py:947: ConvergenceWarning: Liblinear failed to converge, increase the number of iterations.
"the number of iterations.", ConvergenceWarning)
PCA 3D - SVC LINEAR - Test accuracy 0.9272727272727272
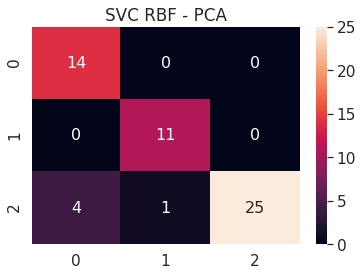
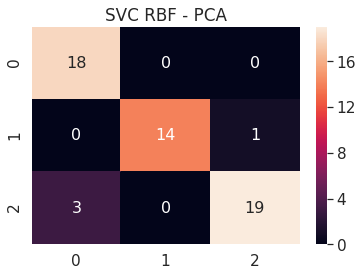
6.A.c SVC RBF - PCA 2D
# SVC RBF PCA 2D
pca_2d_svc_rbf = build_svm(model_type=SVM_MODELS.SVC, x=pca_2d_x_train, y=pca_2d_y_train,
path='models/pca_2d_', svc_kernel='rbf')
print('PCA 2D - SVC RBF - Test accuracy', pca_2d_svc_rbf.score(pca_2d_x_test, pca_2d_y_test))
cm = confusion_matrix(pca_2d_svc_rbf.predict(pca_2d_x_test), pca_2d_y_test)
df_cm = pd.DataFrame(cm, range(n_classes), range(n_classes))
sns.set(font_scale=1.4)
sns.heatmap(df_cm, annot=True, annot_kws={"size": 16})
plt.title('SVC RBF - PCA')
plt.show()
PCA 2D - SVC RBF - Test accuracy 0.9090909090909091
6.A.d SVC RBF - PCA 3D
# SVC RBF PCA 3D
pca_3d_svc_rbf = build_svm(model_type=SVM_MODELS.SVC, x=pca_3d_x_train, y=pca_3d_y_train,
path='models/pca_3d_', svc_kernel='rbf')
print('PCA 3D - SVC RBF - Test accuracy', pca_3d_svc_rbf.score(pca_3d_x_test, pca_3d_y_test))
cm = confusion_matrix(pca_3d_svc_rbf.predict(pca_3d_x_test), pca_3d_y_test)
df_cm = pd.DataFrame(cm, range(n_classes), range(n_classes))
sns.set(font_scale=1.4)
sns.heatmap(df_cm, annot=True, annot_kws={"size": 16})
plt.title('SVC RBF - PCA')
plt.show()
PCA 3D - SVC RBF - Test accuracy 0.9272727272727272
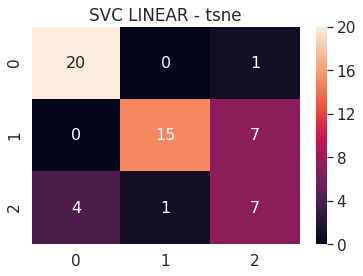
6.B t-SNE
- SVC Linear - t-SNE 2D
- SVC Linear - t-SNE 3D
- SVC RBF - t-SNE 2D
- SVC RBF - t-SNE 3D
6.B.a SVC Linear - t-SNE 2D
# SVC LINEAR TSNE 2D
tsne_2d_svc_linear = build_svm(model_type=SVM_MODELS.LINEAR_SVC, x=tsne_2d_x_train, y=tsne_2d_y_train,
max_iter=200000, path='models/tsne_2d_', multi_class='ovr')
print('tsne 2D - SVC LINEAR - Test accuracy', tsne_2d_svc_linear.score(tsne_2d_x_test, tsne_2d_y_test))
cm = confusion_matrix(tsne_2d_svc_linear.predict(tsne_2d_x_test), tsne_2d_y_test)
df_cm = pd.DataFrame(cm, range(n_classes), range(n_classes))
sns.set(font_scale=1.4)
sns.heatmap(df_cm, annot=True, annot_kws={"size": 16})
plt.title('SVC LINEAR - tsne')
plt.show()
tsne 2D - SVC LINEAR - Test accuracy 0.7636363636363637
6.B.b SVC Linear - t-SNE 3D
# SVC LINEAR TSNE 3D
tsne_3d_svc_linear = build_svm(model_type=SVM_MODELS.LINEAR_SVC, x=tsne_3d_x_train, y=tsne_3d_y_train,
max_iter=200000, path='models/tsne_3d_', multi_class='ovr')
print('tsne 3D - SVC LINEAR - Test accuracy', tsne_3d_svc_linear.score(tsne_3d_x_test, tsne_3d_y_test))
cm = confusion_matrix(tsne_3d_svc_linear.predict(tsne_3d_x_test), tsne_3d_y_test)
df_cm = pd.DataFrame(cm, range(n_classes), range(n_classes))
sns.set(font_scale=1.4)
sns.heatmap(df_cm, annot=True, annot_kws={"size": 16})
plt.title('SVC LINEAR - tsne')
plt.show()
/usr/local/lib/python3.6/dist-packages/sklearn/svm/_base.py:947: ConvergenceWarning: Liblinear failed to converge, increase the number of iterations.
"the number of iterations.", ConvergenceWarning)
tsne 3D - SVC LINEAR - Test accuracy 0.8727272727272727
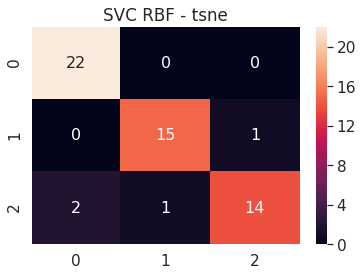
6.B.c SVC RBF - t-SNE 2D
# SVC RBF tsne 2D
tsne_2d_svc_rbf = build_svm(model_type=SVM_MODELS.SVC, x=tsne_2d_x_train, y=tsne_2d_y_train,
path='models/tsne_2d_', svc_kernel='rbf')
print('tsne 2D - SVC RBF - Test accuracy', tsne_2d_svc_rbf.score(tsne_2d_x_test, tsne_2d_y_test))
cm = confusion_matrix(tsne_2d_svc_rbf.predict(tsne_2d_x_test), tsne_2d_y_test)
df_cm = pd.DataFrame(cm, range(n_classes), range(n_classes))
sns.set(font_scale=1.4)
sns.heatmap(df_cm, annot=True, annot_kws={"size": 16})
plt.title('SVC RBF - tsne')
plt.show()
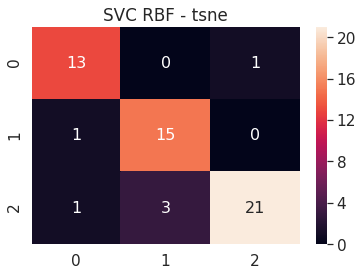
tsne 2D - SVC RBF - Test accuracy 0.9272727272727272
6.B.d SVC RBF - t-SNE 3D
# SVC RBF tsne 3D
tsne_3d_svc_rbf = build_svm(model_type=SVM_MODELS.SVC, x=tsne_3d_x_train, y=tsne_3d_y_train,
path='models/tsne_3d_', svc_kernel='rbf')
print('tsne 3D - SVC RBF - Test accuracy', tsne_3d_svc_rbf.score(tsne_3d_x_test, tsne_3d_y_test))
cm = confusion_matrix(tsne_3d_svc_rbf.predict(tsne_3d_x_test), tsne_3d_y_test)
df_cm = pd.DataFrame(cm, range(n_classes), range(n_classes))
sns.set(font_scale=1.4)
sns.heatmap(df_cm, annot=True, annot_kws={"size": 16})
plt.title('SVC RBF - tsne')
plt.show()
tsne 3D - SVC RBF - Test accuracy 0.8909090909090909
6.C Fully Connected
def fc(input_shape, n_classes, print_summary=True):
inputs = Input(shape=input_shape)
x = Flatten(name='flatten')(inputs)
x = Dense(64, activation='relu', name='fc1')(x)
x = Dense(64, activation='relu', name='fc2')(x)
x = Dense(n_classes, activation='softmax', name='predictions')(x)
model = Model(inputs=inputs, outputs=x)
if print_summary:
model.summary()
return model
fc_model = fc(features.shape[1:], n_classes)
fc_model.compile(loss='categorical_crossentropy', optimizer='adam', metrics=['accuracy'])
import os
def prepare_directory(model_type='vgg19'):
save_dir = os.path.join(os.getcwd(), 'models')
model_name = '%s_model.{epoch:03d}.h5' % model_type
if not os.path.isdir(save_dir):
os.makedirs(save_dir)
filepath = os.path.join(save_dir, model_name)
return filepath
filepath = prepare_directory()
checkpoint = ModelCheckpoint(filepath=filepath, monitor='val_acc', verbose=0, save_best_only=True)
callbacks = [checkpoint]
fc_model.fit(features, y,
validation_split=0.1, epochs=50,
verbose=1, workers=1, callbacks=callbacks)
score = fc_model.evaluate(features, y, batch_size=64)
print(fc_model.metrics_names)
print(score)
Model: "model_5"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
input_8 (InputLayer) (None, 7, 7, 512) 0
_________________________________________________________________
flatten (Flatten) (None, 25088) 0
_________________________________________________________________
fc1 (Dense) (None, 64) 1605696
_________________________________________________________________
fc2 (Dense) (None, 64) 4160
_________________________________________________________________
predictions (Dense) (None, 3) 195
=================================================================
Total params: 1,610,051
Trainable params: 1,610,051
Non-trainable params: 0
_________________________________________________________________
Train on 486 samples, validate on 55 samples
Epoch 1/50
486/486 [==============================] - 1s 1ms/step - loss: 2.8259 - acc: 0.7469 - val_loss: 0.7808 - val_acc: 0.9273
Epoch 2/50
486/486 [==============================] - 0s 229us/step - loss: 0.5015 - acc: 0.9650 - val_loss: 0.6306 - val_acc: 0.9273
Epoch 3/50
486/486 [==============================] - 0s 233us/step - loss: 0.7303 - acc: 0.9403 - val_loss: 0.8186 - val_acc: 0.9091
Epoch 4/50
486/486 [==============================] - 0s 223us/step - loss: 0.3563 - acc: 0.9753 - val_loss: 0.7602 - val_acc: 0.9455
Epoch 5/50
486/486 [==============================] - 0s 236us/step - loss: 0.3539 - acc: 0.9733 - val_loss: 1.4467 - val_acc: 0.8909
Epoch 6/50
486/486 [==============================] - 0s 224us/step - loss: 0.5973 - acc: 0.9609 - val_loss: 0.9528 - val_acc: 0.9091
Epoch 7/50
486/486 [==============================] - 0s 238us/step - loss: 0.4245 - acc: 0.9671 - val_loss: 0.2975 - val_acc: 0.9818
Epoch 8/50
486/486 [==============================] - 0s 242us/step - loss: 0.3037 - acc: 0.9794 - val_loss: 1.2844 - val_acc: 0.9091
Epoch 9/50
486/486 [==============================] - 0s 243us/step - loss: 0.2898 - acc: 0.9774 - val_loss: 0.5861 - val_acc: 0.9636
Epoch 10/50
486/486 [==============================] - 0s 221us/step - loss: 0.2199 - acc: 0.9856 - val_loss: 0.8792 - val_acc: 0.9455
Epoch 11/50
486/486 [==============================] - 0s 218us/step - loss: 0.2319 - acc: 0.9835 - val_loss: 0.8792 - val_acc: 0.9455
Epoch 12/50
486/486 [==============================] - 0s 236us/step - loss: 0.4203 - acc: 0.9691 - val_loss: 0.8792 - val_acc: 0.9455
Epoch 13/50
486/486 [==============================] - 0s 227us/step - loss: 0.2003 - acc: 0.9877 - val_loss: 0.7955 - val_acc: 0.9273
Epoch 14/50
486/486 [==============================] - 0s 224us/step - loss: 0.2901 - acc: 0.9774 - val_loss: 1.5582 - val_acc: 0.8727
Epoch 15/50
486/486 [==============================] - 0s 238us/step - loss: 0.3266 - acc: 0.9774 - val_loss: 0.6782 - val_acc: 0.9455
Epoch 16/50
486/486 [==============================] - 0s 220us/step - loss: 0.1764 - acc: 0.9877 - val_loss: 1.0911 - val_acc: 0.9273
Epoch 17/50
486/486 [==============================] - 0s 222us/step - loss: 0.4236 - acc: 0.9712 - val_loss: 1.4660 - val_acc: 0.9091
Epoch 18/50
486/486 [==============================] - 0s 233us/step - loss: 0.1862 - acc: 0.9856 - val_loss: 1.4653 - val_acc: 0.9091
Epoch 19/50
486/486 [==============================] - 0s 225us/step - loss: 0.1648 - acc: 0.9897 - val_loss: 1.6676 - val_acc: 0.8727
Epoch 20/50
486/486 [==============================] - 0s 216us/step - loss: 0.1048 - acc: 0.9918 - val_loss: 1.1745 - val_acc: 0.9273
Epoch 21/50
486/486 [==============================] - 0s 224us/step - loss: 0.2432 - acc: 0.9815 - val_loss: 0.9253 - val_acc: 0.9273
Epoch 22/50
486/486 [==============================] - 0s 226us/step - loss: 0.1717 - acc: 0.9856 - val_loss: 1.1728 - val_acc: 0.9273
Epoch 23/50
486/486 [==============================] - 0s 232us/step - loss: 0.1682 - acc: 0.9877 - val_loss: 1.3307 - val_acc: 0.9091
Epoch 24/50
486/486 [==============================] - 0s 233us/step - loss: 0.2213 - acc: 0.9835 - val_loss: 2.3918 - val_acc: 0.8364
Epoch 25/50
486/486 [==============================] - 0s 223us/step - loss: 0.4356 - acc: 0.9712 - val_loss: 1.7583 - val_acc: 0.8909
Epoch 26/50
486/486 [==============================] - 0s 221us/step - loss: 0.1415 - acc: 0.9897 - val_loss: 0.5938 - val_acc: 0.9636
Epoch 27/50
486/486 [==============================] - 0s 248us/step - loss: 0.1024 - acc: 0.9918 - val_loss: 0.7616 - val_acc: 0.9455
Epoch 28/50
486/486 [==============================] - 0s 221us/step - loss: 0.1327 - acc: 0.9918 - val_loss: 0.8792 - val_acc: 0.9455
Epoch 29/50
486/486 [==============================] - 0s 214us/step - loss: 0.1327 - acc: 0.9918 - val_loss: 0.8792 - val_acc: 0.9455
Epoch 30/50
486/486 [==============================] - 0s 228us/step - loss: 0.1327 - acc: 0.9918 - val_loss: 0.8792 - val_acc: 0.9455
Epoch 31/50
486/486 [==============================] - 0s 220us/step - loss: 0.1327 - acc: 0.9918 - val_loss: 0.8792 - val_acc: 0.9455
Epoch 32/50
486/486 [==============================] - 0s 232us/step - loss: 0.1327 - acc: 0.9918 - val_loss: 0.8792 - val_acc: 0.9455
Epoch 33/50
486/486 [==============================] - 0s 243us/step - loss: 0.1327 - acc: 0.9918 - val_loss: 0.8792 - val_acc: 0.9455
Epoch 34/50
486/486 [==============================] - 0s 224us/step - loss: 0.1327 - acc: 0.9918 - val_loss: 0.8792 - val_acc: 0.9455
Epoch 35/50
486/486 [==============================] - 0s 221us/step - loss: 0.1327 - acc: 0.9918 - val_loss: 0.8792 - val_acc: 0.9455
Epoch 36/50
486/486 [==============================] - 0s 231us/step - loss: 0.1327 - acc: 0.9918 - val_loss: 0.8792 - val_acc: 0.9455
Epoch 37/50
486/486 [==============================] - 0s 223us/step - loss: 0.1327 - acc: 0.9918 - val_loss: 0.8792 - val_acc: 0.9455
Epoch 38/50
486/486 [==============================] - 0s 224us/step - loss: 0.1327 - acc: 0.9918 - val_loss: 0.8792 - val_acc: 0.9455
Epoch 39/50
486/486 [==============================] - 0s 232us/step - loss: 0.1327 - acc: 0.9918 - val_loss: 0.8792 - val_acc: 0.9455
Epoch 40/50
486/486 [==============================] - 0s 210us/step - loss: 0.1327 - acc: 0.9918 - val_loss: 0.8792 - val_acc: 0.9455
Epoch 41/50
486/486 [==============================] - 0s 213us/step - loss: 0.1327 - acc: 0.9918 - val_loss: 0.8792 - val_acc: 0.9455
Epoch 42/50
486/486 [==============================] - 0s 235us/step - loss: 0.1327 - acc: 0.9918 - val_loss: 0.8792 - val_acc: 0.9455
Epoch 43/50
486/486 [==============================] - 0s 225us/step - loss: 0.1327 - acc: 0.9918 - val_loss: 0.8792 - val_acc: 0.9455
Epoch 44/50
486/486 [==============================] - 0s 225us/step - loss: 0.1327 - acc: 0.9918 - val_loss: 0.8792 - val_acc: 0.9455
Epoch 45/50
486/486 [==============================] - 0s 238us/step - loss: 0.1327 - acc: 0.9918 - val_loss: 0.8792 - val_acc: 0.9455
Epoch 46/50
486/486 [==============================] - 0s 220us/step - loss: 0.1327 - acc: 0.9918 - val_loss: 0.8792 - val_acc: 0.9455
Epoch 47/50
486/486 [==============================] - 0s 210us/step - loss: 0.1327 - acc: 0.9918 - val_loss: 0.8792 - val_acc: 0.9455
Epoch 48/50
486/486 [==============================] - 0s 224us/step - loss: 0.1327 - acc: 0.9918 - val_loss: 0.8792 - val_acc: 0.9455
Epoch 49/50
486/486 [==============================] - 0s 218us/step - loss: 0.1327 - acc: 0.9918 - val_loss: 0.8792 - val_acc: 0.9455
Epoch 50/50
486/486 [==============================] - 0s 223us/step - loss: 0.1327 - acc: 0.9918 - val_loss: 0.8792 - val_acc: 0.9455
541/541 [==============================] - 0s 74us/step
['loss', 'acc']
[0.20855218491581207, 0.9870609971599966]
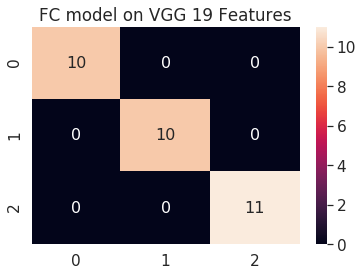
print('VGG 19 - FC - Test accuracy', fc_model.evaluate(features_test, y_test, batch_size)[1])
cm = confusion_matrix(np.argmax(fc_model.predict(features_test), axis=1), np.argmax(y_test, axis=1))
df_cm = pd.DataFrame(cm, range(n_classes), range(n_classes))
sns.set(font_scale=1.4)
sns.heatmap(df_cm, annot=True, annot_kws={"size": 16})
plt.title('FC model on VGG 19 Features')
plt.show()
31/31 [==============================] - 0s 252us/step
VGG 19 - FC - Test accuracy 1.0
 Nikan
Nikan  Imbalance Learning and Evalution using AdaBoost, RUSBoost, SMOTEBoost, RBBoost and ANOVA Measure
Imbalance Learning and Evalution using AdaBoost, RUSBoost, SMOTEBoost, RBBoost and ANOVA Measure